Photo Competition finished and we have a winner!
The winner is Renping Zhao, project A2
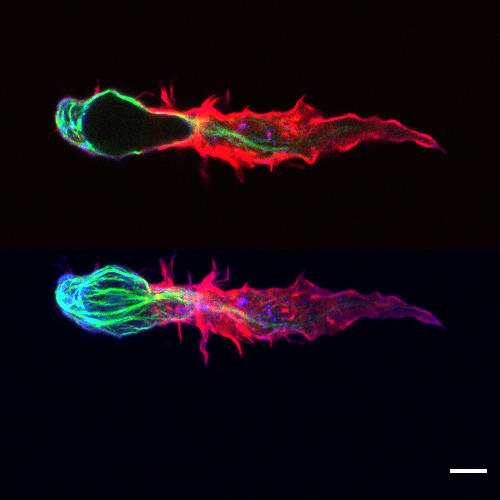
Fig.1 - Morphology of primary human CD8+ T cell in 3D matrix In project A2, we focus on T cell function in 3D environment. A migrating primary human CD8+ T cell was fixed in 3D matrix of 4 mg/ml bovine collagen I. Microtubule was labeled with microtubule binding domain of ensconsin (EMTB) -3XGFP (green); F-actin was labeled with Alex 568-phalloidin (red); and CD3 was labeled with Alexa 647 anti-human CD3 antibody (blue). Image was acquired with LSM confocal microscopy. The upper panel is single layer of z-stack images and the lower one is the maximum intensity projection of the z-stacks. Scale bar: 5 µm.
The second place is shared by Friederike Nolle and Thomas Faidt, project B2
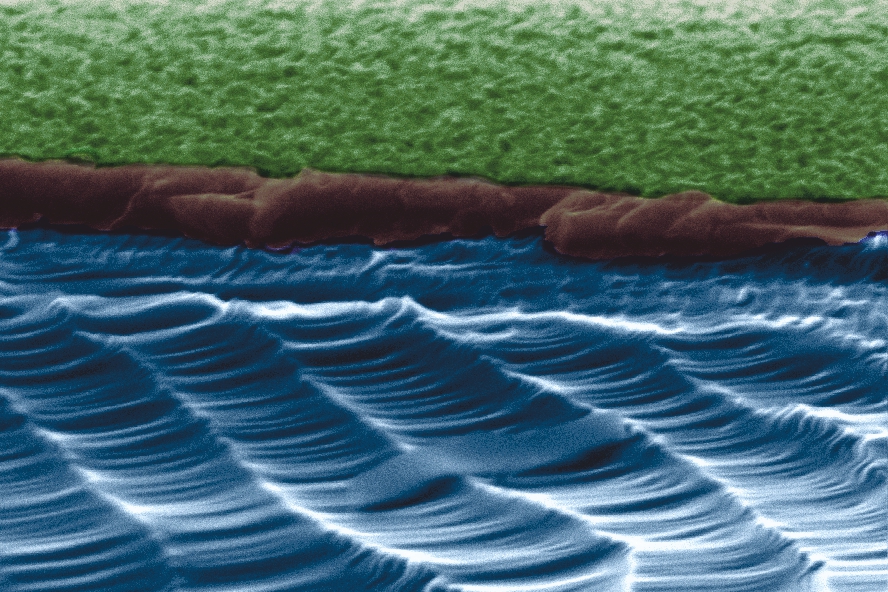
Fig.2 - Scanning electron microscopy image of a wet chemical etched silicon surface (Rq: 7.5 nm) at a breaking edge, also called Black Silicon. The scanning electron microscopy images were take at the INM with help of Dr. Koch. In project B2, we study the adhesion process of pathogenic bacteria. The ability of pathogenic bacteria to great biofilms on various surfaces is a nuisance in a wide area of health care applications. We could show that this nano-rough surface doesn't affect the adhesion of Staphylococcus aureus cells strongly as the roughness is too small.
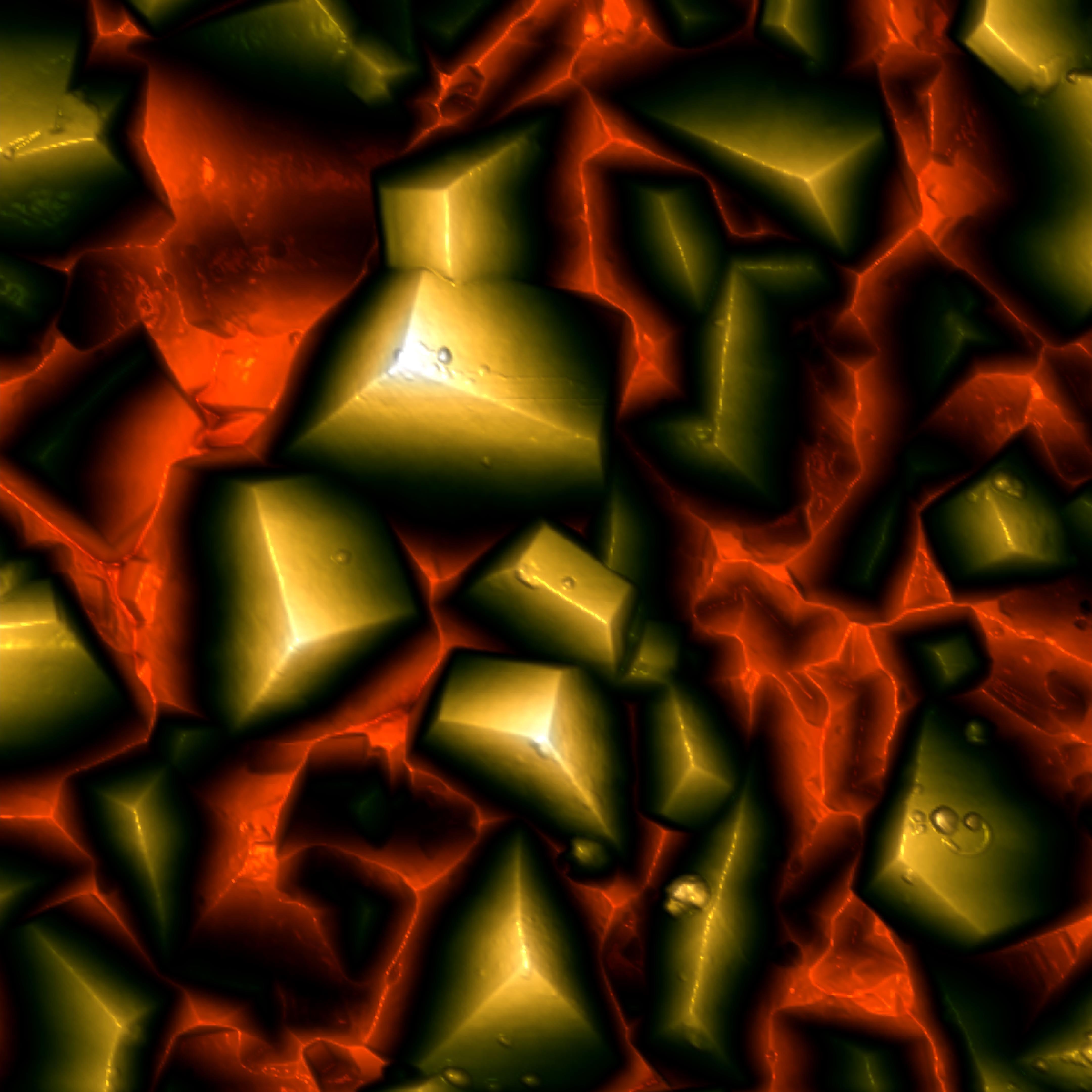
Fig.3 - 10 x 10 µm² AFM height scan of µm-sized diamonds grown on a Si(001)-surface for 16 hours. The height scale from red to white is 1 µm. In project B1, we use tailored surfaces to investigate interactions between, e.g., proteins and substrates. One of these tailored surfaces is hydroxyapatite, the main component of tooth enamel. For our experiments the surfaces need to be extremely smooth. Therefore, we tried to use rough diamond surfaces grown under different conditions as 'sandpaper' to polish the hydroxyapatite surface. The diamond crystals have been grown by the 'AG Diamant' at Uni Augsburg.
The third place belongs to Zahra Mostajeran, project A9
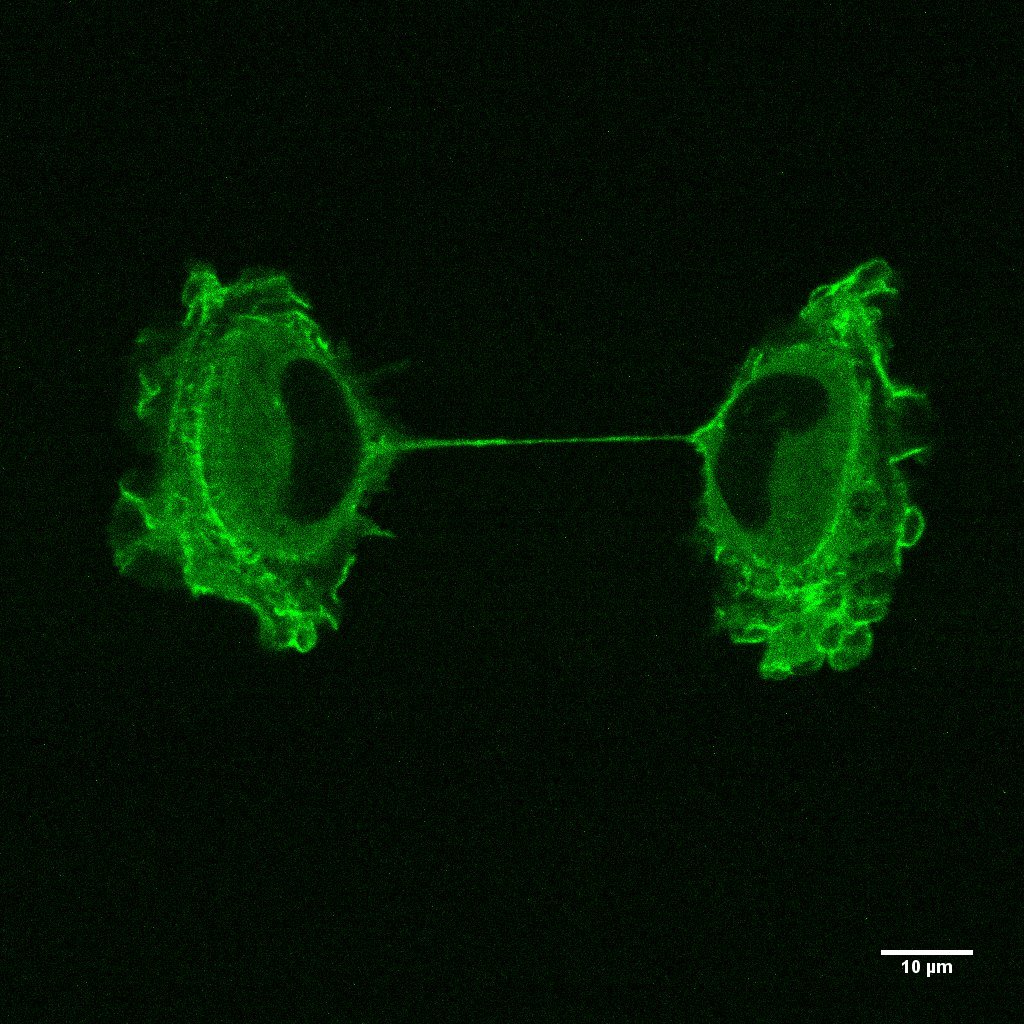
Fig.4 - RPE1 cells were labeled by Actin GFP BacMam technology (using an insect cell virus (baculovirus)), the sample is imaging by confocal laser scanning microscopy in different Z levels in the last step of mitosis (Cytokinesis). Z ≈10um We label actin filaments in living cells by Actin GFP BacMam to investigate Actin dynamics in cell cortex by fluorescence recovery after photo-bleaching technology and study cell cortex structure during force generation.


